Transduction of E. coli CDC11-1 with P1 Lysates
A Summary of E. coli Transduction with P1
November 4th, 2021Introduction
Previously in the Biotechnology lab, we have prepared phage lysate solutions from two E. coli strains (JW5055 & JW0391) using the P1 virus with the supervision of Dr. Deutch. Currently, our goal is to carry out transduction of these lysate solutions into the triple mutant E. coli strain CDC11-1 (delta PutP, delta ProP, and delta ProW). Transduction is the process where a selected gene of interest can be integrated into a new bacterial cell using a phage. When the bacteriophage P1kc was introduced to the strains JW5055 (a strain that is delta ProY::kan) and JW0391 (a strain that is delta BrnQ::kan), the lytic cycle of the virus production resulted in some mutations that lead to the packaging of bacterial DNA along with viral DNA inside the phage head. We were hopeful that the ProY::kan and the BrnQ::kan deletion genes were packaged into some P1kc viruses to help carry out our transduction procedure. The hypothesis of this experiment is whether using different concentrations of the P1(JW5055) and P1(JW0391) phage suspensions would produce different transduction results (different numbers of colonies) when these virus lysates are mixed with the E. coli strain CDC11-1.
Procedure
Dr. Deutch grew 5 ml culture of E. coli CDC11-1 in LB broth at home which we will be using as the target of transduction. The procedure starts by centrifuging 1.5 ml portions of CDC11-1 in two microcentrifuge tubes to separate the concentrated cells from the medium. The centrifugation is done at 10,000 rpm for 5 minutes. After the medium is discarded into a waste beaker, 750 microliters of P1 MC salts solution containing magnesium sulfate and calcium chloride are added to and mixed with both microcentrifuge tubes containing CDC11-1 cells. The MC salts are essential for viral attachment to host cells. The content of the tubes is then divided into sterile 13 x 100 tubes labeled 1 through 8. In each tube, a 100 microliters of E. coli CDC11-1 with MC solution are added, then an amount of phage suspension is determined for addition. Tubes 1 and 5 are the control tubes, so no phage lysates are added to them. Tubes 2-4 are for the transduction with P1(JW5055), and tubes 6-8 are for the transduction with P1(JW0391). Amounts of 2 microliters, 10 microliters and 50 microliters of each P1 lysate solution will be used in corresponding tubes. The tables for this information is the following:
After mixing the solutions with the corresponding phage suspensions, a 20 minutes waiting period follows to allow the phages to attach to new bacterial cells and infect them with the DNA they carry, both viral and bacterial. After the waiting period, virus attachment needs to be blocked by a solution containing sodium citrate to allow for the DNA to divide and gene expression inside the host cells to take place. Thus, 8 ml of LB broth is mixed with 2 ml of 1.0 M sodium citrate to then be added to the solutions made prior containing the phage suspensions. That is, 1.0 ml of the LB + sodium citrate is added to each of the 8 tubes and the mixtures are then incubated at 37 degrees Celsius for an hour. A step of centrifugation follows after the contents are transferred into corresponding microcentrifuge tubes labeled 1 to 8. After about 5 minutes of centrifugation, we end up with the pellet on the bottom of the microcentrifuge tubes and the medium on top. The medium is then discarded and each pellet is resuspended in 100 microliters of the same solution of LB + sodium citrate to ensure the absolute blockage of transduction. The suspension of each tube is then plated onto a LB with kanamycin Petri dish labeled with the same number as the tube using a sterilized hockey stick. The last step includes incubating the 8 Petri dishes at 37 degrees Celsius for two days.
Results and Conclusion
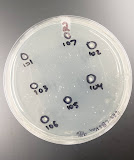
After the two days incubation period, the 8 Petri dishes are brought out for analyzation. The number of colonies on each plate is counted. Results are shown below:
*Plate four had no growth, so it is not shown above.
We expected no growth in the control plates with no phage suspension added, which are plates one and five; however, growth was present which suggests that there was contamination during the process of transduction. Plate one had 10 colonies while plate five had around 13 colonies. Due to this, the transduction procedure will be repeated next week. Moreover, plate two had 7 colonies, plate three had 1 colony, plates six and seven had 10 colonies, and plate eight had 1 colony. Plates 2-4 were made with the lysate from P1(JW5055), so the corresponding colonies may have the ProY::kan gene integrated in their DNA. Plates 6-8 were made with the lysate from P1(JW0391), so their colonies may have the BrnQ::kan gene in their DNA as well. This explains the ability of the new quadruple mutant CDC11-1, previously kanamycin sensitive and now delta ProY or delta BrnQ, to grow on kanamycin due to it becoming kanamycin resistant from the transduced mutations by the P1kc virus. The results also confirm that the higher the amount of the phage suspension, the less colonies present. For example, plate two had only 2 microliters of the phage lysate while plate three had 10 microliters of the lysate; thus, plate two shows more numerous colonies than plate three, confirming the hypothesis. These results are consistent with plates 6-8 as well. To elaborate, a higher amount of the virus lysate means that more viruses will lyse more bacterial cells, leaving a less concentrated bacterial culture; therefore, colonies appear less numerous than those with a lower amount of virus lysate added. The photos above show labeled colonies for a future application which would involve the purification of these transductants on fresh LB + kanamycin plates.
References
Deutch, C.E. (October 2021) Transduction of E. coli CDC11-1 with P1(JW5055) and P1(JW0391). Put Project at GCC.







Comments
Post a Comment